× close
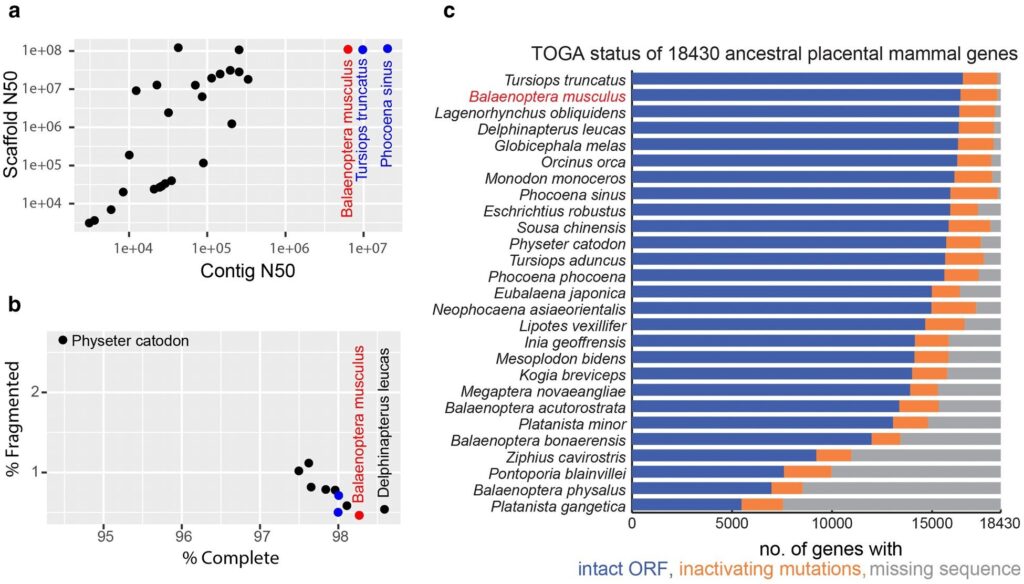
Assembly quality indicators. Blue whale (Balaenoptera musculus) data is displayed in red. His other two VGP assemblies, the porpoise (Phocoena sinus) and the bottlenose dolphin (Tursiops truncatus), are blue. a) Assembly contig and scaffold N50 metrics. Contigs are contiguous, or gapless, segments of sequence. A scaffold is a set of contigs that are ordered and oriented using optical maps or long-range mapping data such as Hi-C with gaps between contigs. N50 is a measure of average length, for example, 50% of all bases are found in contigs with length N50 or longer. b) Proportion of complete and fragmented universal single-copy BUSCO orthologs found within the annotated genome. A universal single-copy ortholog is a gene that is present in a single copy in all or most genomes within a phylogenetic group. A high % complete score indicates that the genome assembly is not missing large amounts of genetic coding sequences (Simão et al. 2015; Manni et al. 2021). C) TOGA status of 18,430 ancestral placental mammalian genes. Note: Different assemblies were used in panel C compared to panel A for the two species. GCA_004363415.1 was used instead of GCA_002189225.1 for Eschrichtius robotus, and GCA_008795845.1 was used instead of GCA_023338255.1 for Balaenoptera physalus. credit: molecular biology and evolution (2024). DOI: 10.1093/molbev/msae036
of blue whale genome It was published in the magazine molecular biology and evolutionand the Etruscan shrew genome It was published in the magazine scientific data.
Research models using animal cell cultures can help answer big biological questions, but these tools are only useful if you follow the right maps.
“The genome is the blueprint of an organism,” said Yury, first author of the published study and a computational biologist in the Ron Stewart Computational Group at the Morgridge Institute, an independent research institute working in conjunction with the University of Wisconsin.・Mr. Bukman says: Madison researches emerging fields such as regenerative biology, metabolism, virology, and biomedical imaging.
“You need to know the genome of that species in order to manipulate cell cultures and measure things like gene expression. That allows you to do more research.”
The Morgridge team’s interest in blue whales and Etruscan shrews stems from the development of the developmental clock by James Thomson, Director Emeritus of Regenerative Biology at Morgridge University and longtime professor of cell and regenerative biology at the University of California School of Medicine. It began with research into the biological mechanisms behind it. and public health.
It is generally known that large organisms take longer to grow from fertilized eggs to adults than smaller organisms, but the reason for this is not yet understood.
“From that perspective, just the basic biological knowledge is important: How are such large animals made? How do they function?” Buchman says.
Buchmann suggests that a practical application of this knowledge lies in the emerging field of stem cell-based therapies. To heal an injury, stem cells must differentiate into specialized cell types in the relevant organ or tissue. The rate of this process is controlled by some of the same molecular mechanisms underlying the developmental clock.
What the genomes of animals of different sizes can tell us about our own health
Understanding the genomes of the largest and smallest mammals may also help solve a biomedical puzzle known as the Peto paradox. This is because large mammals such as whales and elephants live longer and are less likely to develop cancer, even though they have more cells (and therefore more cell division). This is an interesting phenomenon. It is often caused by occasional DNA replication errors during cell division. ) than humans and small mammals such as mice.
Meanwhile, knowledge of the Etruscan shrew genome will enable new insights in the field of metabolism. Shrews have a very high surface area to volume ratio and a fast metabolic rate. These high energy demands are a product of their small size, no larger than a human thumb and weighing less than a penny, making them an interesting model for better understanding metabolic regulation.
The Blue Whale and Etruscan Shrew Genome Project, in conjunction with the Vertebrate Genome Project, is part of a larger collaboration involving dozens of contributors from institutions in several countries in North America and Europe.
VGP’s mission is to collect high-quality reference genomes for all extant vertebrate species on Earth. This international consortium of researchers includes top experts in genome assembly and curation.
“VGP has established a set of methods and standards for creating reference genomes,” Buchmann says. “Accuracy, continuity, and completeness are his three measures of quality.”
For more information:
Yury V Bukhman et al. High-quality blue whale genomes, segmental duplications, and historical demography, molecular biology and evolution (2024). DOI: 10.1093/molbev/msae036
Yury V. Bukhman et al, Chromosome-level genome assembly of the Etruscan shrew Suncus etruscus, scientific data (2024). DOI: 10.1038/s41597-024-03011-x